Árboles de decisión¶
Los árboles de decisión son un método de aprendizaje supervisado no paramétrico utilizado para la clasificación y la regresión, es un modelo que no es sensible a los NA ni a valores atipicos.
los árboles de decisión aprenden de los datos para aproximar curva con un conjunto de reglas de decisión if/else, ver ejemplo acontinuación.
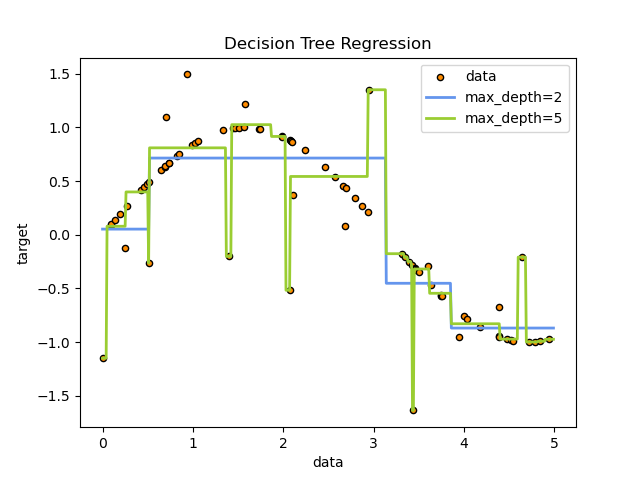
Un arbol de decisión posee los siguientes elementos:
- nodo raiz
- ramas
- nodos de decisión
- nodo hojas
# Cargando librerias
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
# leyendo la base de datos del repositorio de unalytics
data = pd.read_csv("https://raw.githubusercontent.com/unalyticsteam/databases/master/iris.csv")
data.head(3)
Preparando los datos.¶
Lo primero es identificar las variables que me serviran de predictores y cual la varible objetivo.
# tomando los nombres de las variables en una lista
colnames = data.columns.values.tolist()
# identificando predictores y variable de respuesta
predictors = colnames[:4]
target = colnames[4]
Tomemos una muestra aleatoria del aproximadamente el 75 % de las observaciones como datos de entrenamiento y el restante 25 % como datos de prueba.
# creando una columna para identificar cuales observaciones nos serviran como datos de prueba.
data["is_train"] = np.random.uniform(low=0, high=1, size= len(data)) <= 0.75
data.head(5)
# tomando los datos de entrenamiento y los datos de prueba
train, test = data[data["is_train"] == True] , data[data["is_train"]== False]
# observemos las primeras entradas
train.head(3)
# Importando librerias
# sklearn: biblioteca para aprendizaje de máquina de software libre (Incluye varios algoritmos de clasificación, regresión y análisis de grupos)
from sklearn.tree import DecisionTreeClassifier
# creando el árbol.
tree = DecisionTreeClassifier(criterion="entropy", min_samples_split=20, random_state= 99)
tree.fit(train[predictors], train[target])
Predicción¶
# predicciones
preds = tree.predict(test[predictors])
# Matriz de confusión.
pd.crosstab(test[target], preds, rownames=["acutual"], colnames=["predictors"])
Vusualización¶
# Instalacion en colab.
! pip install graphviz
# importando libreria de visualización.
from sklearn.tree import export_graphviz
# hay que exportar como .dot (grafos)
with open("dtree.dot", "w") as dotfile:
export_graphviz(tree, out_file=dotfile, feature_names=predictors)
dotfile.close()
# leyendo el objeto.
from graphviz import Source
file = open("dtree.dot", "r")
text = file.read()
Source(text)
Validación cruzada.¶
X = data[predictors]
Y = data[target]
tree = DecisionTreeClassifier(criterion="entropy", max_depth= 5,
min_samples_split=20, random_state=99)
tree.fit(X, Y)
from sklearn.model_selection import KFold
cv = KFold(X.shape[0], shuffle=True, random_state=1 )
from sklearn.model_selection import cross_val_score
scores = cross_val_score(tree, X, Y, scoring="accuracy", cv = cv, n_jobs=1)
scores
score = np.mean(scores)
score
Árboles de Regresión¶
# Impotando librerias
import pandas as pd
from sklearn.tree import DecisionTreeRegressor
Leyendo los datos de las casas de Boston(1978), del repositorio de unalytics
data = pd.read_csv("https://raw.githubusercontent.com/unalyticsteam/databases/master/Boston.csv")
crim: indice de criminologia per-capita.(100 induviduos)
zn: proporcion de zona de residencia por cada pie cudrado
indus: Acceso a zona de industria.
chas: cerca rio de boston (0, 1)
rm: promedio de habitaciones.
...
Rápido Análisis¶
# Observemos las 3 primeras filas
data.head(3)
observemos las variables, tipo de dato, memoria.
# dimensión de la base de datos.
data.shape
# informacion de la base de datos.
data.info()
Pre-Procesando base de datos.¶
colnames = data.columns.values.tolist()
colnames
# preprocesado
predictors = colnames[0:13]
targets = colnames[13]
x = data[predictors]
y = data[targets]
regtree = DecisionTreeRegressor(min_samples_split=30, min_samples_leaf=10, random_state=0)
regtree.fit(x,y) #predictoras, prediccion
preds = regtree.predict(data[predictors])
data["preds"] = preds
data[["preds", "medv"]].head()
Visualizacion¶
from sklearn.tree import export_graphviz
from sklearn.tree import export_graphviz
with open("boston_rtree.dot", "w") as dotfile:
export_graphviz(regtree, out_file=dotfile, feature_names=predictors)
dotfile.close()
import os
from graphviz import Source
file = open("boston_rtree.dot", "r")
text = file.read()
Source(text)
